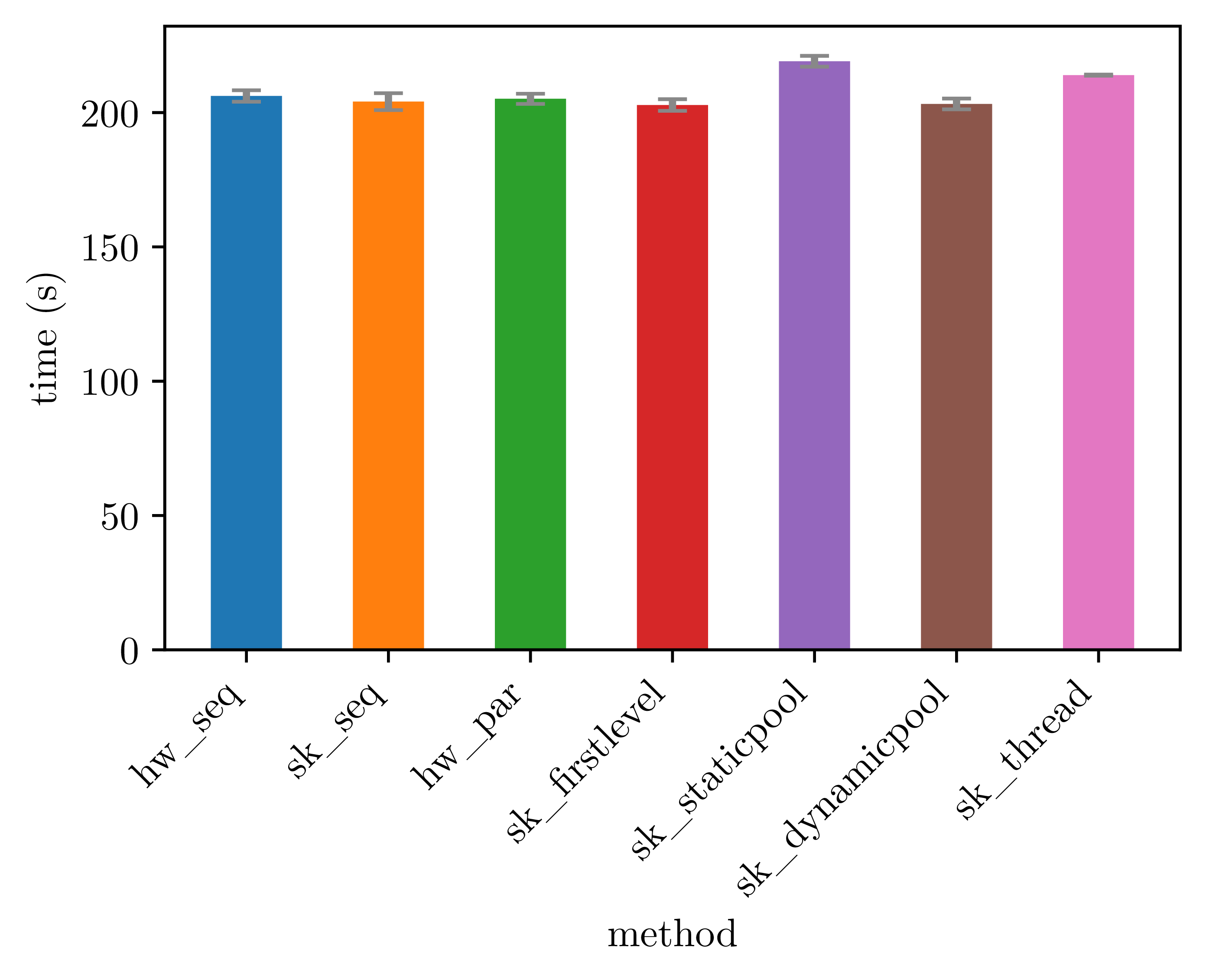About
ROSA (in french "Recherche Opérationnelle grâce aux Squelettes Algorithmiques", i.e. Operational Research with Algorithmic Skeletons). It relies on alsk for the algorithmic skeletons. This is part of the work done for my Ph.D. thesis.
Brief
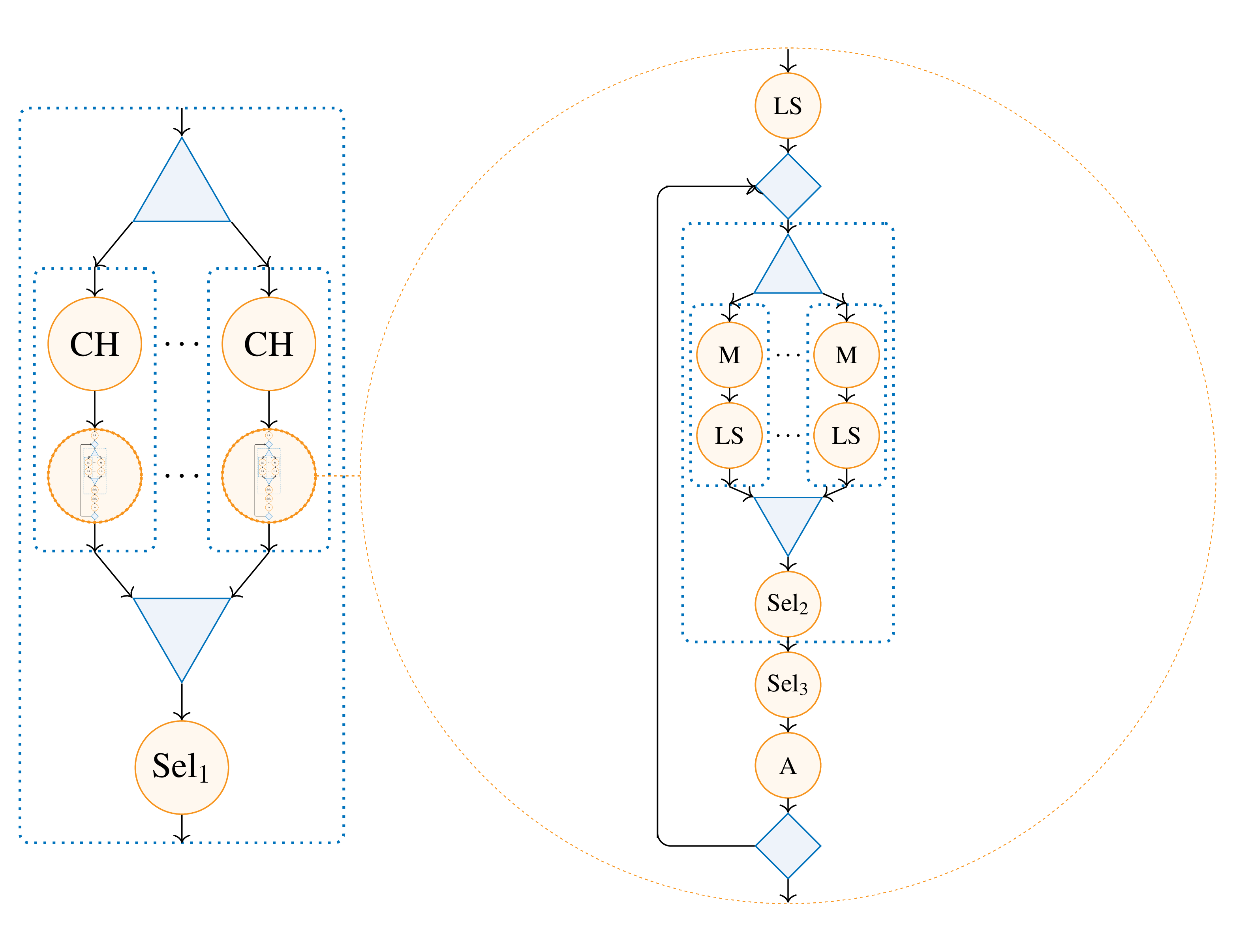
The main algorithm implemented and presented is GRASP×ELS. This metaheuristic can be represented as below:
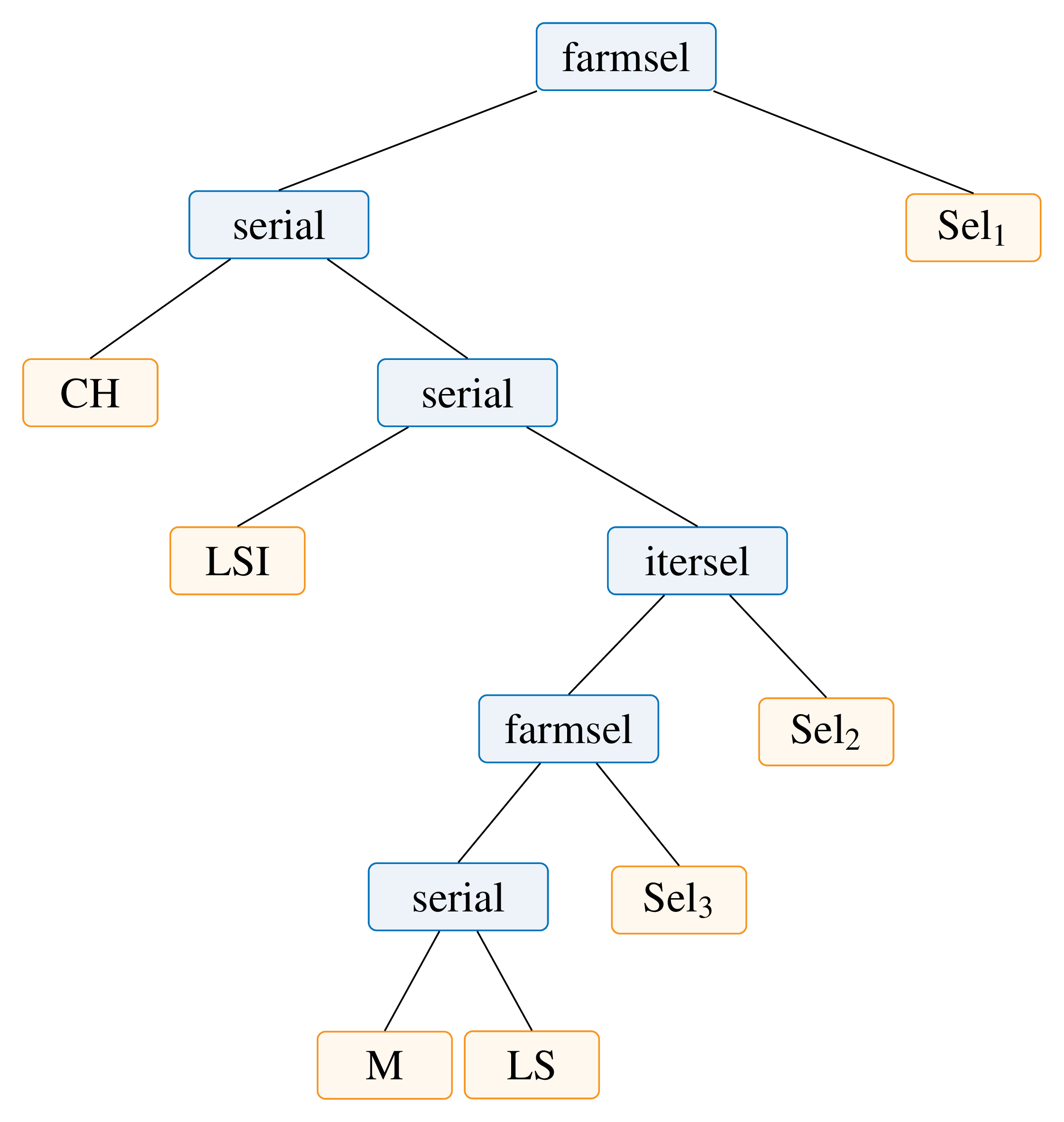
When implemented as an algorithmic skeleton, its representation becomes this tree:
Obtained using the following source code.
For the internal ELS:
template<typename InitLS, typename Mutate, typename LS, typename InnerSelect, typename OuterSelect>
using SkelElsStruct =
S<alsk::Serial,
InitLS, // LSI
S<IterSel,
S<FarmSel,
S<Serial,
Mutate, LS // M then LS
>,
InnerSelect // Sel3
>,
OuterSelect // Sel2
>
>;
template<typename Solution>
using SkelElsLinks =
L<Serial, R<1>(Solution const&),
Solution(P<0>),
L<IterSel, Solution(R<0> const&),
L<FarmSel, Solution(Solution),
L<Serial, R<1>(P<0>),
Solution(P<0>, RNG&),
Solution(R<0> const&)
>,
Solution(Solution const&, Solution const&)
>,
Solution(Solution const&, Solution const&)
>
>;
template<
typename Solution,
typename InitLS, typename Mutate, typename LS, typename InnerSelect, typename OuterSelect
>
using SkelEls = BuildSkeleton<SkelElsStruct, SkelElsLinks>::skeleton<
Pack<InitLS, Mutate, LS, InnerSelect, OuterSelect>,
Pack<Solution>
>;
For the GRASP:
template<typename CH, typename LS, typename Select>
using SkelGraspStructure =
S<FarmSel,
S<Serial, CH, LS>,
Select // Sel1
>;
template<typename Problem, typename Solution>
using SkelGraspLinks =
L<FarmSel, Solution(Problem const&),
L<Serial, R<1>(P<0>),
Solution(P<0>, RNG),
Solution(R<0>)
>,
Solution(Solution, Solution)
>;
template<typename Problem, typename Solution, typename CH, typename LS, typename Select>
using SkelGrasp = BuildSkeleton<SkelGraspStructure, SkelGraspLinks>::skeleton<
Pack<CH, LS, Select>,
Pack<Problem, Solution>
>;
Then the GRASP×ELS can be constructed:
// All arguments are defined types or functions, see full source code
using ELS = SkelEls<
tsp::Solution,
Descent,
Move2Opt, Descent,
FN(selectMin)
>;
using GRASPxELS = SkelGrasp<
tsp::Problem, tsp::Solution,
RGreedy<tsp::Solution>, ELS,
FN(selectMin)
>;
Performances
The measures shown below are from using the GRASPxELS algorithm to solve an instance of TSP with 194 nodes. Various execution policies are used:
- "hw_seq": handwritten sequential implementation;
- "hw_par": handwritten parallel implementation;
- "sk_seq": skeleton without parallelisation;
- "sk_firstlevel": skeleton with parallelisation of the first level;
- "sk_staticpool": skeleton with parallelisation using a thread pool with static task distribution;
- "sk_dynamicpool": skeleton with parallelisation using a classical thread pool with dynamic task distribution;
- "sk_thread": skeleton with parallelisation using dynamically created threads.
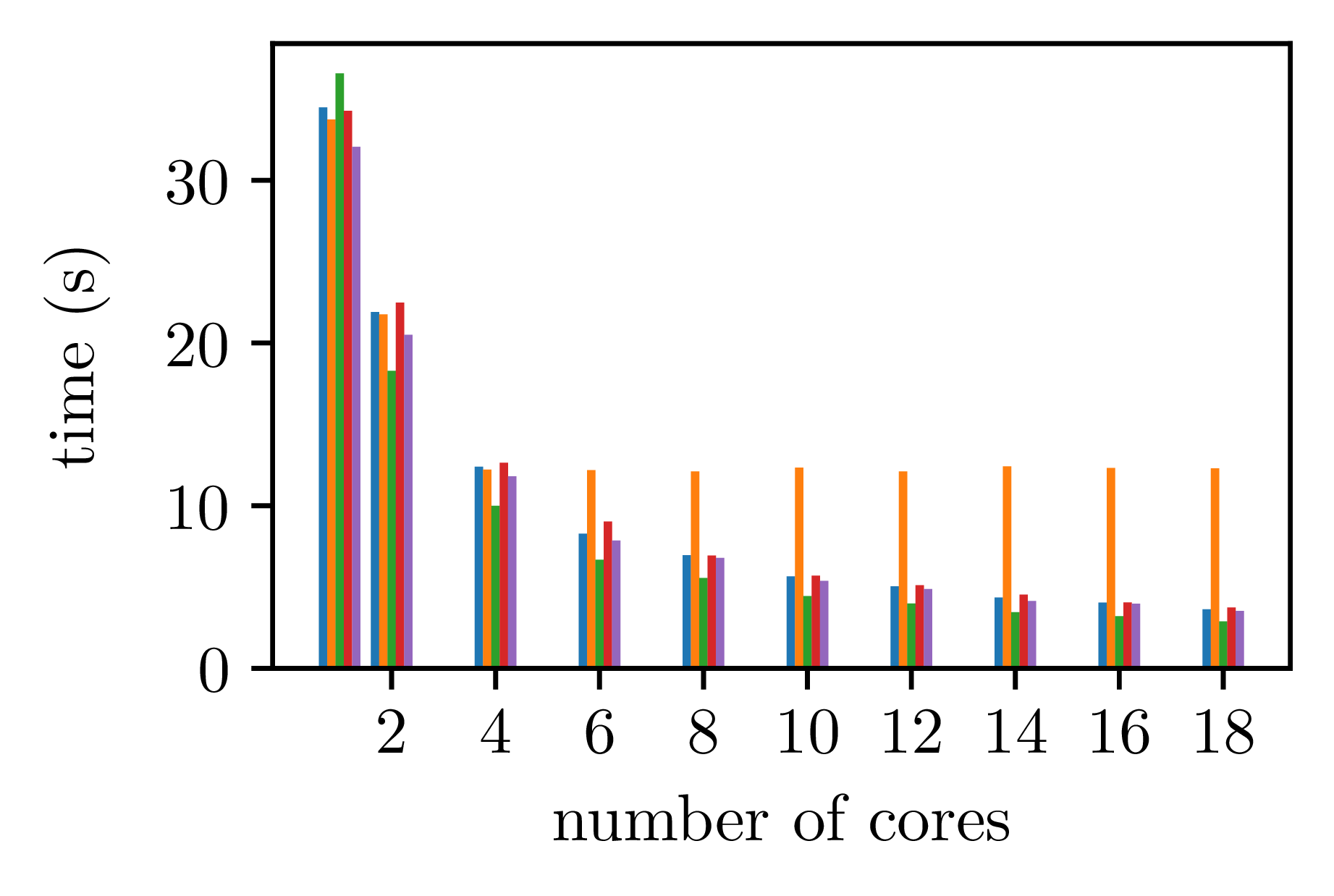
For an execution with only one allotted core, meaning that there is no parallelisation done, we obtain the data below. Note that this data set do not use the legend shown above. All subsequent images use it.
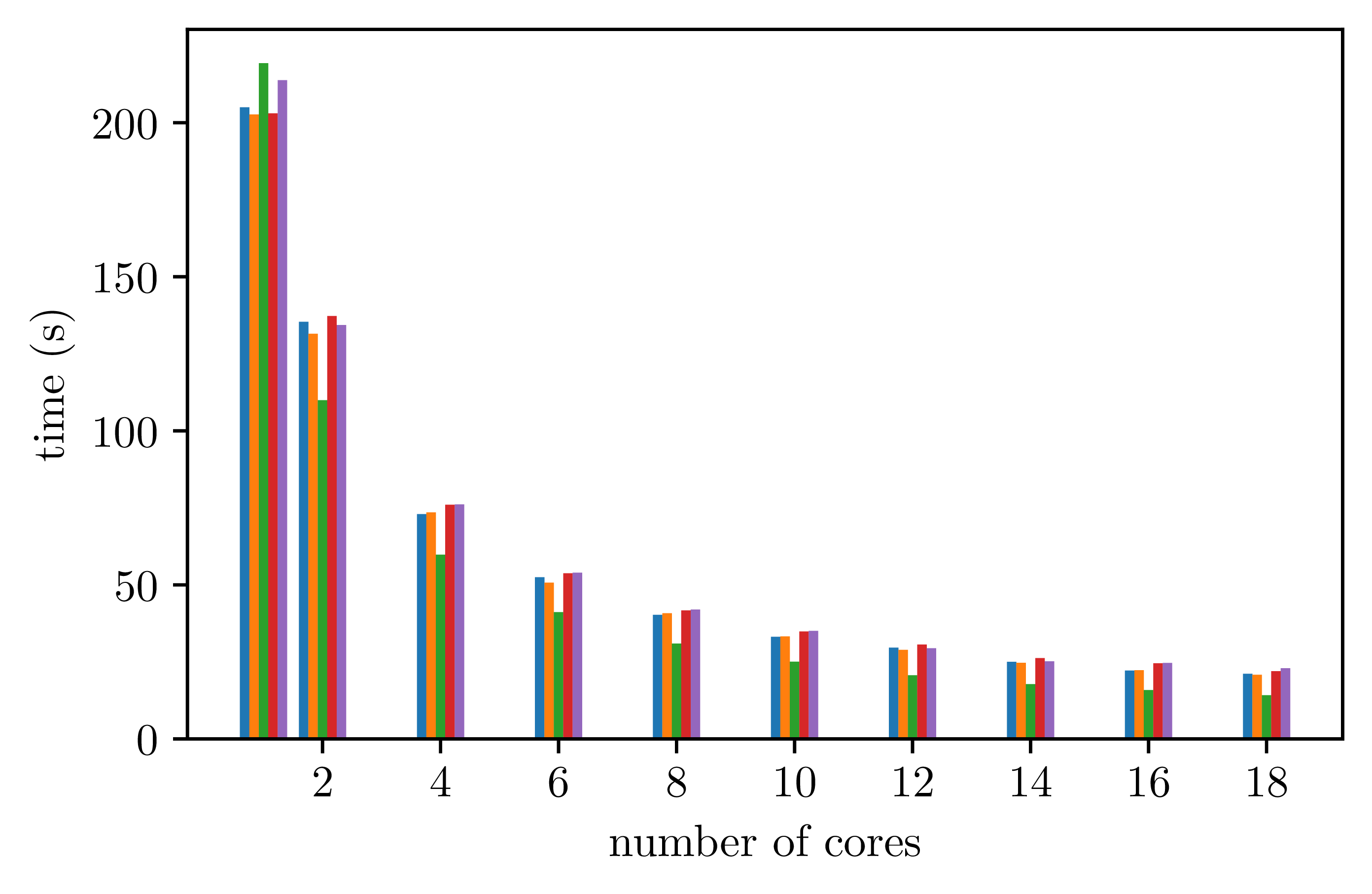
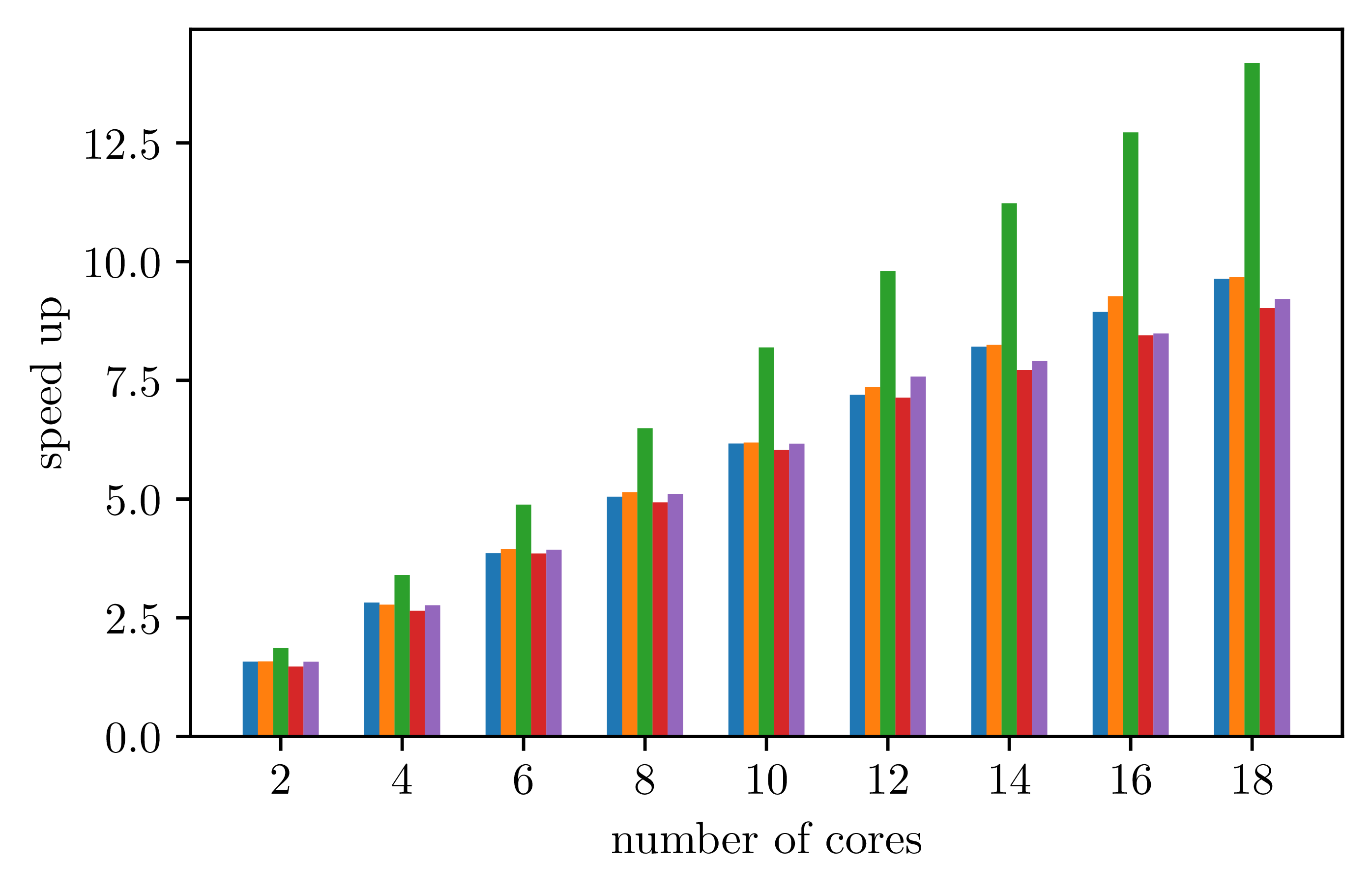
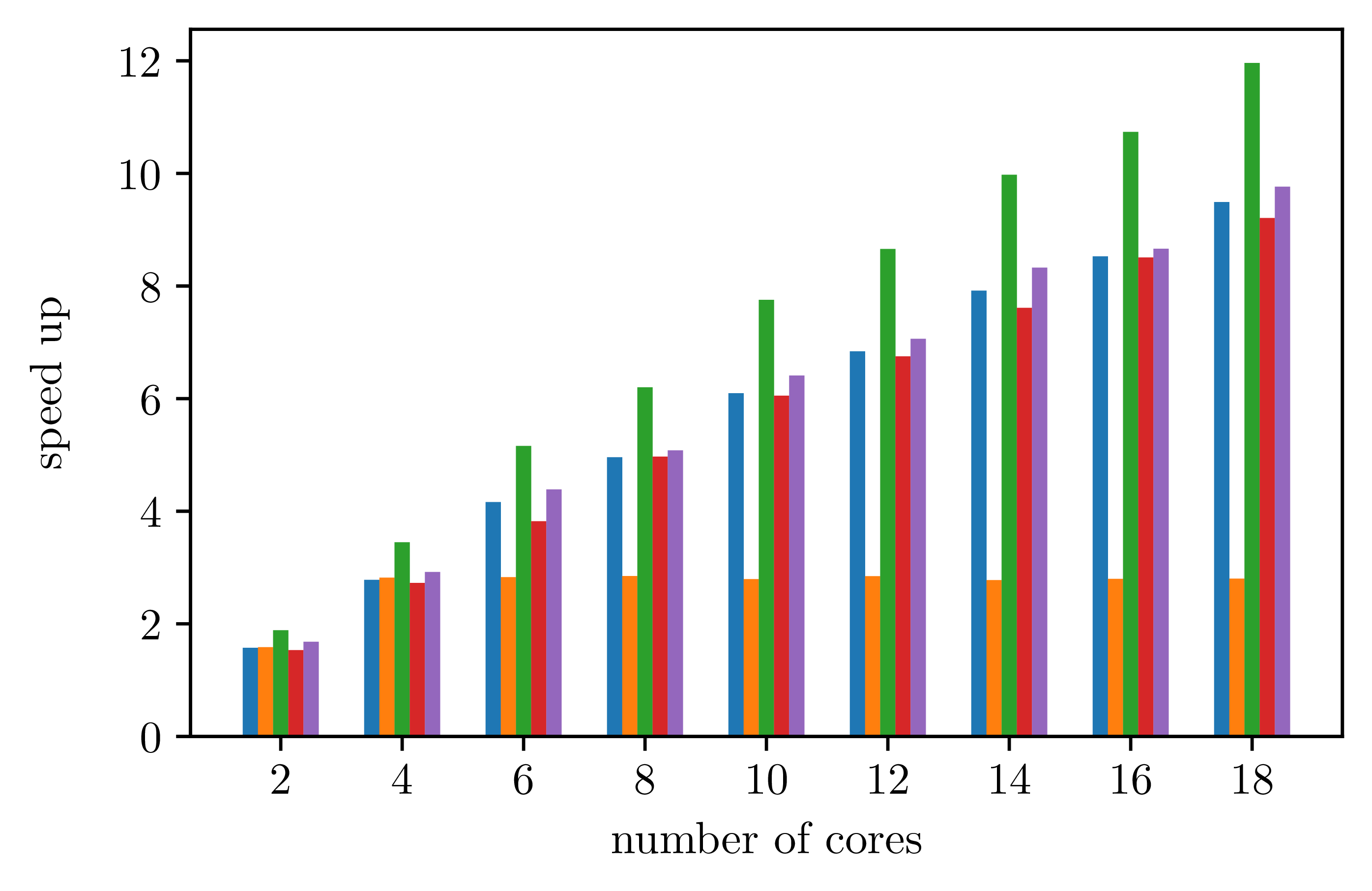
For parallel executions, measures give the following data.
With 24 iterations for the outmost parallel loop:
With only 4 iterations for the outmost parallel loop:
Related publications
- "Repeatability with Random Numbers Using Algorithmic Skeletons", ESM 2020 (https://hal.archives-ouvertes.fr/hal-02980472);
- "Modeling Algorithmic Skeletons for Automatic Parallelization Using Template Metaprogramming", HPCS 2019 (IEEE) 10.1109/HPCS48598.2019.9188128;
- "Processing Algorithmic Skeletons at Compile-Time", ROADEF 2020 (https://hal.archives-ouvertes.fr/hal-02573660);
- "Algorithmic Skeletons Using Template Metaprogramming", ICAST 2019;
- "Parallel Algorithmic Skeletons for Metaheuristics", ROADEF 2019 (https://hal.archives-ouvertes.fr/hal-02059533).
Organisation
Main directories:
src: sources;src/rosa: the library sources;rtbenchmarks: scripts for compile-time/run-time benchmarking;results: results presented in the thesis, obtained using mentioned scripts and codes.
Usage
To produce the Makefile and build the project:
mkdir build
cd build
cmake -DCMAKE_BUILD_TYPE=Release ..
make